PARTICIPATING LABS
Suter, David
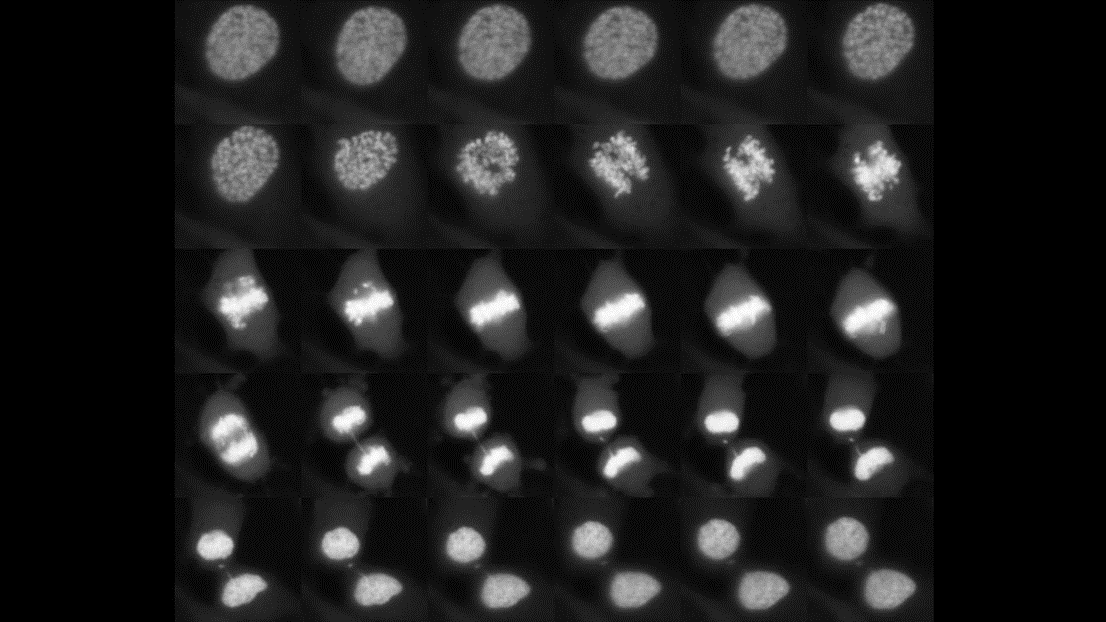
Our laboratory is focused on the control of cell identity by mechanisms acting throughout the gene expression cascade, from transcription to protein degradation. To do so, we develop and leverage quantitative approaches of gene expression in stem cells and cancer cells to dissect gene regulation at the single cell level and throughout the cell cycle. More specifically, we perform quantitative analysis of DNA-binding proteins and histones using single cell and single molecule approaches, to understand how their activity depends on their biophysical properties, temporal fluctuations of their expression levels, and interplay with the cell cycle. We have also developed approaches allowing lineage tracing based on gene expression memory, and quantitative monitoring of transcription & protein turnover in single living cells. We combine these live cell technologies with genome-wide analysis of transcription factor binding (ChIP-seq, CUT&TAG), chromatin accessibility and nucleosome positioning (ATAC-seq, MNase-seq) transcription (RNA-seq/single cell RNA-seq).
Key technologies
- Genomics (ChIP-seq
- CUT&TAG, ATAC-seq
- RNA-seq (bulk & single cell)
- single cell multiomics, MNase-seq)
- Proteomics
- Quantitative live cell imaging
- & computational inference of live cell trajectories
- Live cell single molecule imaging
- Stem cell differentiation
Key biological questions
- How are dynamic properties of chromatin and transcription factors controlling self-renewal ?
- How are cells regulating their proteome concentration in health and disease ?
Contact
EPFL SV IBI-SV UPSUTERAI 1241 (Bâtiment AI)
Station 19
1015 Lausanne





