PARTICIPATING LABS
Bastings, Maartje
The Programmable Biomaterials Laboratory (PBL) is part of the Institute of Materials (IMX) and the interfaculty bioengineering institute (IBI) at EPFL. We are experts in DNA-nanotechnology, supramolecular self-assembly and cellular particle uptake and stability. We explore how precise spatial organization of molecules can control multivalent binding behavior between soft-matter and cells in a dynamic reciprocal fashion.
The goal of the Programmable Biomaterials Lab is to integrate dynamic reciprocity, e.g. a two-way action-reaction feedback loop, into supramolecular nanomaterials. Exploiting the multitude of dynamic non-covalent interactions presented by DNA, we want to create materials that self-organize into functional architectures, dictated by a dynamic interplay with cellular interactions. Herefore, we need to understand how spatial organization of active components and material geometry can be used as parameters to predict and control hierarchical self-organization and emergent properties of surfaces, in particles and in soft matter. Combining materials engineering with biophysics and immune biology, we lift DNA to become a quantifiable biomaterial with unprecedented control in cellular (inter)action.
Our research towards dynamic multivalency in DNA-based nanomaterials focuses on:
1) Dynamic materials with emergent structure and function through programmable control of self-organization
2) Design of novel binding behavior through the engineering of spatial tolerance in multivalent interactions
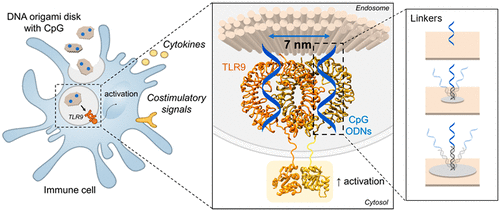
3) Precise manipulation of immune signaling through material geometry and control of functionality
Key technologies
- DNA nanotechnology
- cell uptake
- nanomaterials
- ligand-receptor interactions
- targeting
- multivalency
Key biological questions
- How do cells use the spatio-temporal clustering of receptors to control signalling in immune activation and cancer?
Contact
EPFL - STI - IMX - PBL (Office MXC340)Station 12
CH-1015 Lausanne, Switzerland





