PARTICIPATING LABS
Bucher, Philipp
Multi-omics molecular profiling assays have produced vast amounts of public data, which could potentially tell us a lot about the processes that cause a healthy cell to become cancerous. The two main axes of our research are:
- Development of new computational resources to make sense out of data
- Biological question-driven analysis of genomics and epigenomics data
Regarding Cancer, we have participated in three computational challenges to predict tumor phenotypes from flow cytometry, proteomics and transcriptomics data, respectively. We have further developed computational methods for making better use of tumor RNA-seq data, including one for somatic mutation calling and one for cross-study standardization. Current work focuses on epigenetics data analysis of breast cancer samples. In collaboration with Cathrin Brisken’s group we are developing the Breast Cancer Epigenomics Track Hub, a platform for visualization, integration and analysis of public ChIP-seq data, with a focus on DNA elements and proteins involved in hormone receptor signaling.
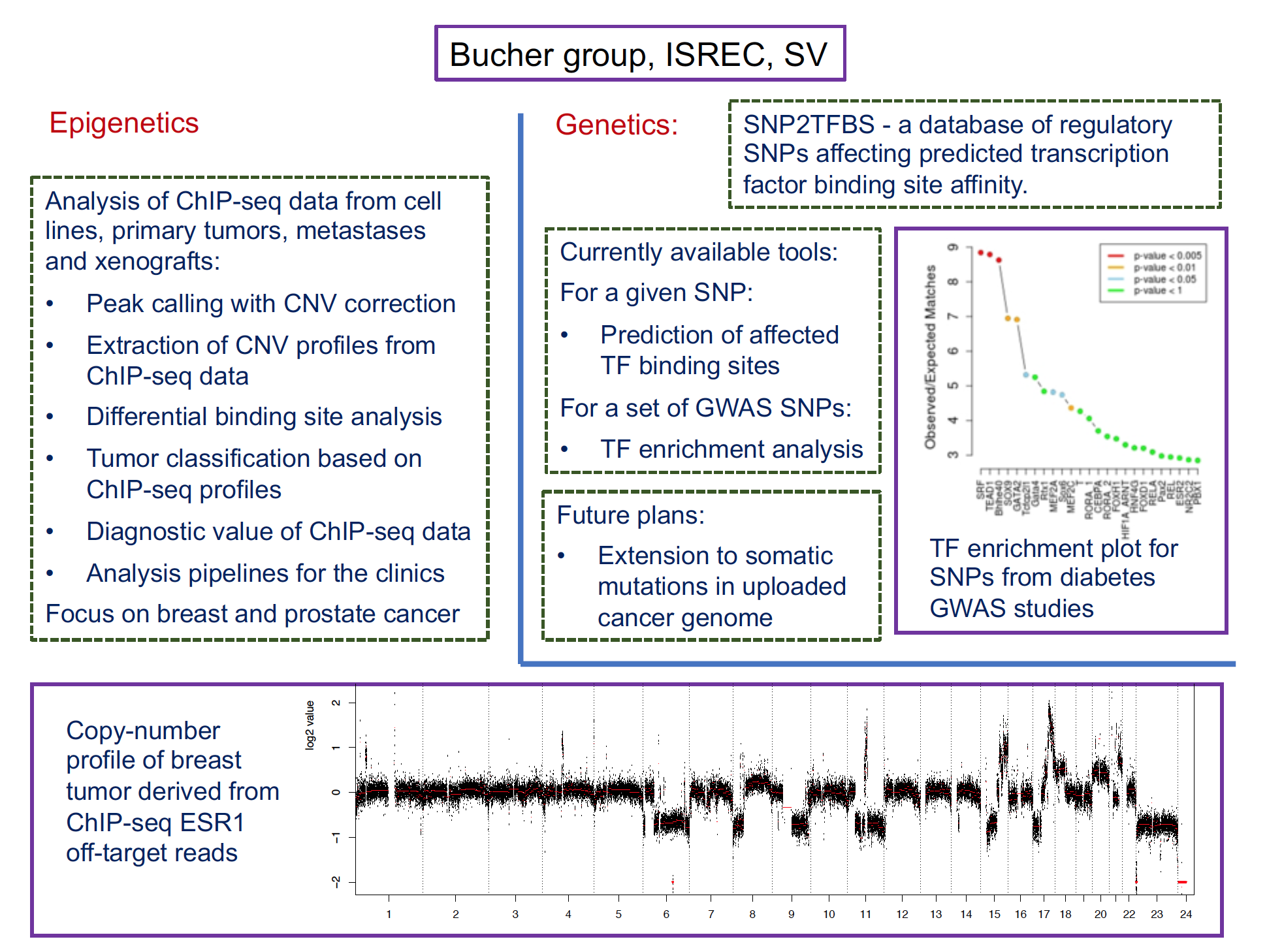
Key technologies
- Genomics data analysis and visualization
- Machine-learning applications to clinical data
- Algorithm and software development
- Web-based bioinformatics resources
- Data curation and database design
Key biological questions
- How useful are epigenetic data for tumor classification, prognosis and prediction?
- Which high-throughput technologies are most cost-effective for personalized medicine?
- What are the challenges and limitations in epigenetic data analysis of tumor samples and how can we overcome them?
- What is the potential of liquid biopsies combined with omics assays in the field of cancer diagnosis?
- How can we make relevant pieces of data hidden in large public omics data sets findable and useable to bench biologists interested only in one ore a few genes?
Contact
EPFL SV SSV ENSAAB 0 17
Station 19
CH-1015 Lausanne
Focus areas
Tumor Evolution
Cutting Edge Diagnosis





